
The research team: Juli Feigon, Yaqiang Wang, Jiansen Jiang, Lukas Sušac and Z. Hong Zhou (left to right) in front of the California NanoSystems Institute at UCLA. Photo courtesy of UCLA Newsroom.
Adapted from UCLA Newsroom article
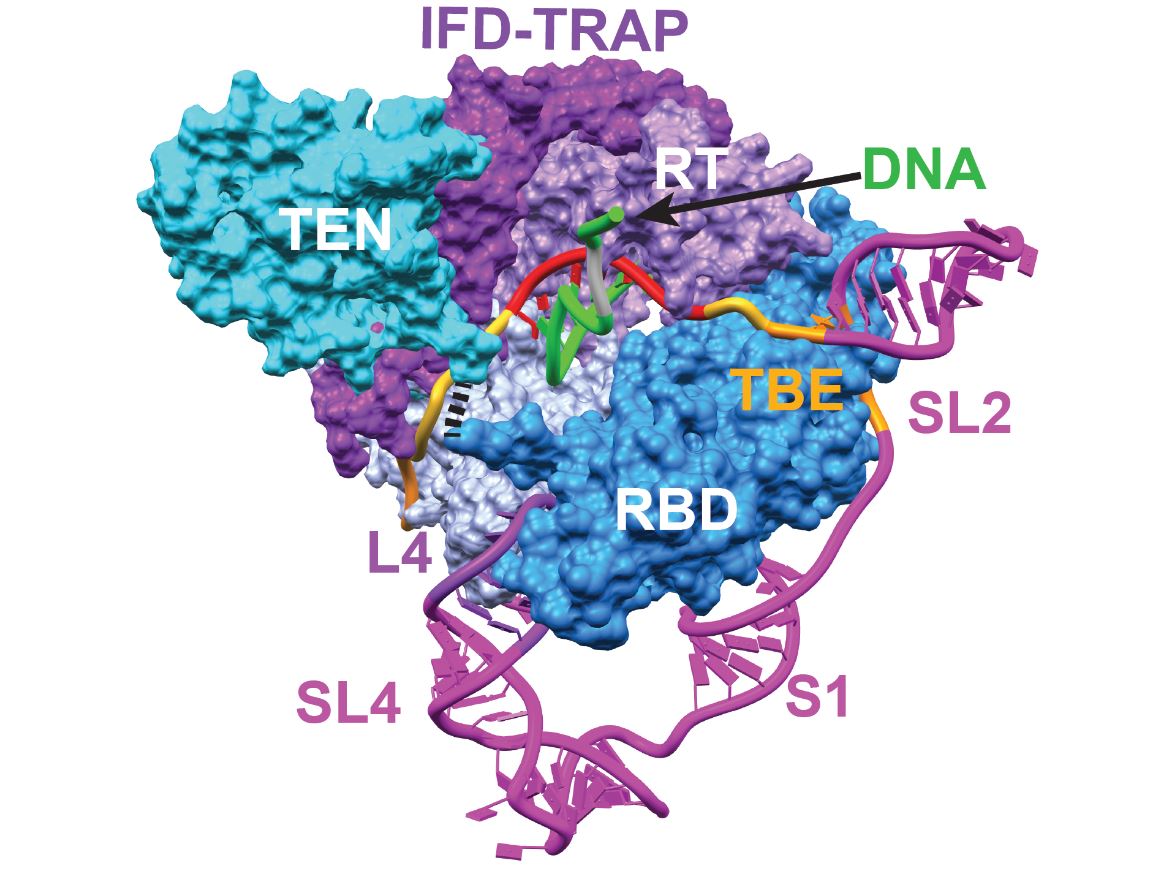
Efforts to combat the symptoms of aging and diseases such as cancer have been underway for almost a century. However, researchers have been unable to target the sources of these complex human health issues on a deep, molecular level. Dr. Juli Feigon, Distinguished Professor of Chemistry and Biochemistry at UCLA and Dr. Hong Zhou, Professor of Microbiology, Immunology, and Molecular Genetics, Director of the California NanoSystems Institute and a Committee Member at the Institute for Digital Research and Education (IDRE) at UCLA, collaborated to find the missing piece to this puzzle. Detailed in their report in the journal Cell titled “Structure of Telomerase with Telomeric DNA”, they constructed three-dimensional maps of Tetrahymena telomerase, an RNA-protein complex that maintains the ends of chromosomes. Recent advancements in technology have allowed the researchers to determine the structure of this enzyme, which is a critical determinant of aging, cancer cell multiplication, and stem cell renewal. For the first time, the scientists were able to view telomerase in near-atomic resolution, this is the highest level of detail that researchers have been able to study the enzyme at, and have even captured telomerase in the process of making DNA. Read more about the results.

Telomerase’s catalytic core. Photo courtesy of Juli Feigon, et al./UCLA/Cell.
Using a technique called cryoEM (cryo electron microscopy) the scientists are able to collect the data needed to create a three-dimensional (3D) map of telomerase and other molecular complexes not visible by conventional methods. In order to achieve this, “processing the large amount of data needed for atomic resolution reconstructions requires either accessing to very expensive computer clusters or waiting for weeks of continuous computation in a personal computer (PC)”.2 To help minimize wait time and resources, scientists calculated portions of the research using graphics processing units (GPUs) on PCs. The use of GPUs generated the same data and accuracy needed to create 3D models. GPUs provide a less expensive alternative and reduced time from weeks–months to only hours–days for high resolution 3D reconstructions of large complexes. Read more about atomic resolution cryoEM reconstruction.
Authors included in “Structure of Telomerase with Telomeric DNA” are Dr. Juli Feigon, senior author and lead contact, Dr. Z. Hong Zhou senior author, Dr. Jiansen Jiang co-lead author, Dr. Yaqiang Wang co-lead author, Dr. Lukas Sušac co-lead author, Dr. Ritwika Basu, and Dr. Henry Chan. Read more about the research in the UCLA Newsroom article.
Citations:
- Jiang J, Wang Y, Sušac L, Chan H, Basu R, Zhou ZH, Feigon J. Structure of Telomerase with Telomeric DNA. Cell. 2018 May 17;173(5):1179-1190.e13. doi: 10.1016/j.cell.2018.04.038. PubMed PMID: 29775593; PubMed Central PMCID: PMC5995583.
- Zhang, Xiaokang, Xing Zhang, and Z. Hong Zhou. “Low Cost, High Performance GPU Computing Solution for Atomic Resolution CryoEM Single-Particle Reconstruction.” Journal of Structural Biology 172.3 (2010): 400–406. PMC. Web. 20 July 2018. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3382114/
- “DNA Replication: Telomeres and telomerase”. Khan Academy. 23 July 2018. https://www.khanacademy.org/science/biology/dna-as-the-genetic-material/dna-replication/a/telomeres-telomerase
- Wolpert, Stuart. “Scientists see inner workings of enzyme telomerase, which plays key roles in aging, cancer”. UCLA Newsroom. 6 June 2018.http://newsroom.ucla.edu/releases/scientists-see-inner-workings-of-enzyme-telomerase-which-plays-key-roles-in-aging-cancer